Build Template Database
In this notebook, we use the events detected in notebook 5 to build a database of template events. This step consists mostly in building data and metadata files but also allows the selection of events based on quality and the grouping of similar events. We want to avoid having too many similar templates in the database as it means useless, redundant computation in the template matching process (similar templates detect the same events).
[1]:
import BPMF
import copy
import glob
import os
import sys
import h5py as h5
import matplotlib.pyplot as plt
import numpy as np
from time import time as give_time
from tqdm import tqdm
from BPMF.data_reader_examples import data_reader_mseed
from multiprocessing import cpu_count
n_CPUs = max(cpu_count(), 24)
os.environ["OMP_NUM_THREADS"] = str(n_CPUs)
[19]:
%config InlineBackend.figure_formats = ['svg']
[2]:
# program parameters
DB_FILENAME = "output_bp_tuto.h5"
DB_PATH_T = "template_db"
DATA_FOLDER = "preprocessed_2_12"
NETWORK_FILENAME = "network.csv"
[3]:
# read network metadata
net = BPMF.dataset.Network(NETWORK_FILENAME)
net.read()
Read the database of detected events
We read the database of events detected with backprojection and, with them, create BPMF.dataset.Template instances.
[4]:
templates = []
tid = 0
with h5.File(os.path.join(BPMF.cfg.OUTPUT_PATH, DB_FILENAME), mode="r") as f:
print(f"Checking {DB_FILENAME}...")
for key in tqdm(f.keys()):
event = BPMF.dataset.Event.read_from_file(
DB_FILENAME, db_path=BPMF.cfg.OUTPUT_PATH, gid=key
)
if "NLLoc_reloc" in event.aux_data:
if event.hmax_unc < 10.0:
# if event was well relocated with NLLoc, use its location
event.set_moveouts_to_theoretical_times()
event.read_waveforms(
BPMF.cfg.TEMPLATE_LEN_SEC,
data_folder=DATA_FOLDER,
data_reader=data_reader_mseed,
)
template = BPMF.dataset.Template.init_from_event(event)
template.id = tid
template.set_aux_data({"tid": tid})
templates.append(template)
tid += 1
Checking output_bp_tuto.h5...
100%|██████████| 13/13 [00:02<00:00, 4.81it/s]
Remove redundant templates
We first create an BPMF.dataset.TemplateGroup instance with the templates built in the previous cell. This class offers a number of methods to operate on groups of templates.
[5]:
template_group = BPMF.dataset.TemplateGroup(templates, net)
# this trick is necessary to make the class aware that the waveforms are already
# attached to the Template instances (_update_attributes should never be called at
# any other time of the workflow).
template_group._update_attributes.append('read_waveforms')
# normalize the waveforms
template_group.normalize()
ERROR 1: PROJ: proj_create_from_database: Open of /home/ebeauce/miniconda3/envs/hy7_py310/share/proj failed
[6]:
# parameters controlling how the inter-template correlation matrix is computed
DISTANCE_THRESHOLD_KM = 5. # in km, criterion on ellipsoid separation
N_CLOSEST_STATIONS = 8 # > larger than total number of stations, i.e. use all stations
MAX_LAG_SAMP = 10 # in samples, maximum time shift allowed when searching for similarity
TemplateGroup.compute_intertemplate_cc computes the network-averaged correlation coefficient between templates. To account for possible location errors – and, therefore, different alignments on the P- and S-wave arrivals – we search for the maximum correlation within +/- MAX_LAG_SAMP.
[7]:
t1 = give_time()
template_group.compute_intertemplate_cc(
distance_threshold=DISTANCE_THRESHOLD_KM,
n_stations=N_CLOSEST_STATIONS,
save_cc=False,
max_lag=MAX_LAG_SAMP,
compute_from_scratch=True,
)
t2 = give_time()
print(f"Computed inter-template CCs in {t2-t1:.2f}sec.")
Read inter-template CCs from ../BPMF_data/2012/intertp_cc.h5.
Computing the similarity matrix...
Computing the inter-template directional errors...
/home/ebeauce/miniconda3/envs/hy7_py310/lib/python3.10/site-packages/BPMF/dataset.py:2832: RuntimeWarning: invalid value encountered in true_divide
unit_direction /= np.sqrt(np.sum(unit_direction**2, axis=1))[
Computed inter-template CCs in 3.11sec.
[8]:
# set the threshold to consider two template as redundant
# this parameter is quite arbitrary and really depends on how much computation
# time you are ready to spend in the matched-filter search
# templates with very low correlation coefficients (~0.10) still detect
# many events in common
SIMILARITY_THRESHOLD = 0.50
[9]:
templates_filtered = []
tids_processed = []
for tid in template_group.tids:
if tid in tids_processed:
continue
tt = template_group.tindexes.loc[tid]
similar = template_group.intertemplate_cc.loc[tid] > SIMILARITY_THRESHOLD
n_similar = np.sum(similar)
if n_similar > 1:
similar_tids = template_group.intertemplate_cc[similar].index
print(f"Template {tid} is similar to: {similar_tids}")
tids_processed.extend(similar_tids.tolist())
best_tid = -10
best_unc = 10000000.0
for tid_sim in similar_tids:
tt_sim = template_group.tindexes.loc[tid_sim]
unc = np.sqrt(
template_group.templates[tt_sim].hmax_unc ** 2
+ template_group.templates[tt_sim].vmax_unc ** 2
)
if unc < best_unc:
best_unc = unc
best_tid = tid_sim
print(f"Keeping only Template {best_tid}")
templates_filtered.append(
template_group.templates[
template_group.tindexes.loc[best_tid]
]
)
elif n_similar == 0:
print(
f"Problem with Template {tid}, it should at least be similar with" " itself"
)
else:
tids_processed.append(tid)
templates_filtered.append(template_group.templates[tt])
Template 0 is similar to: Int64Index([0, 1, 3, 5, 9], dtype='int64')
Keeping only Template 0
Template 6 is similar to: Int64Index([6, 8], dtype='int64')
Keeping only Template 8
Template 7 is similar to: Int64Index([3, 5, 7], dtype='int64')
Class instance does not have a `cov_mat` attribute.
Keeping only Template 5
Plot the similar templates
How good was our criterion to group similar templates together? Let’s check the waveforms of the similar templates.
[11]:
fig = template_group.templates[template_group.tindexes.loc[0]].plot()
fig.suptitle(f"Template 0")
[11]:
Text(0.5, 0.98, 'Template 0')
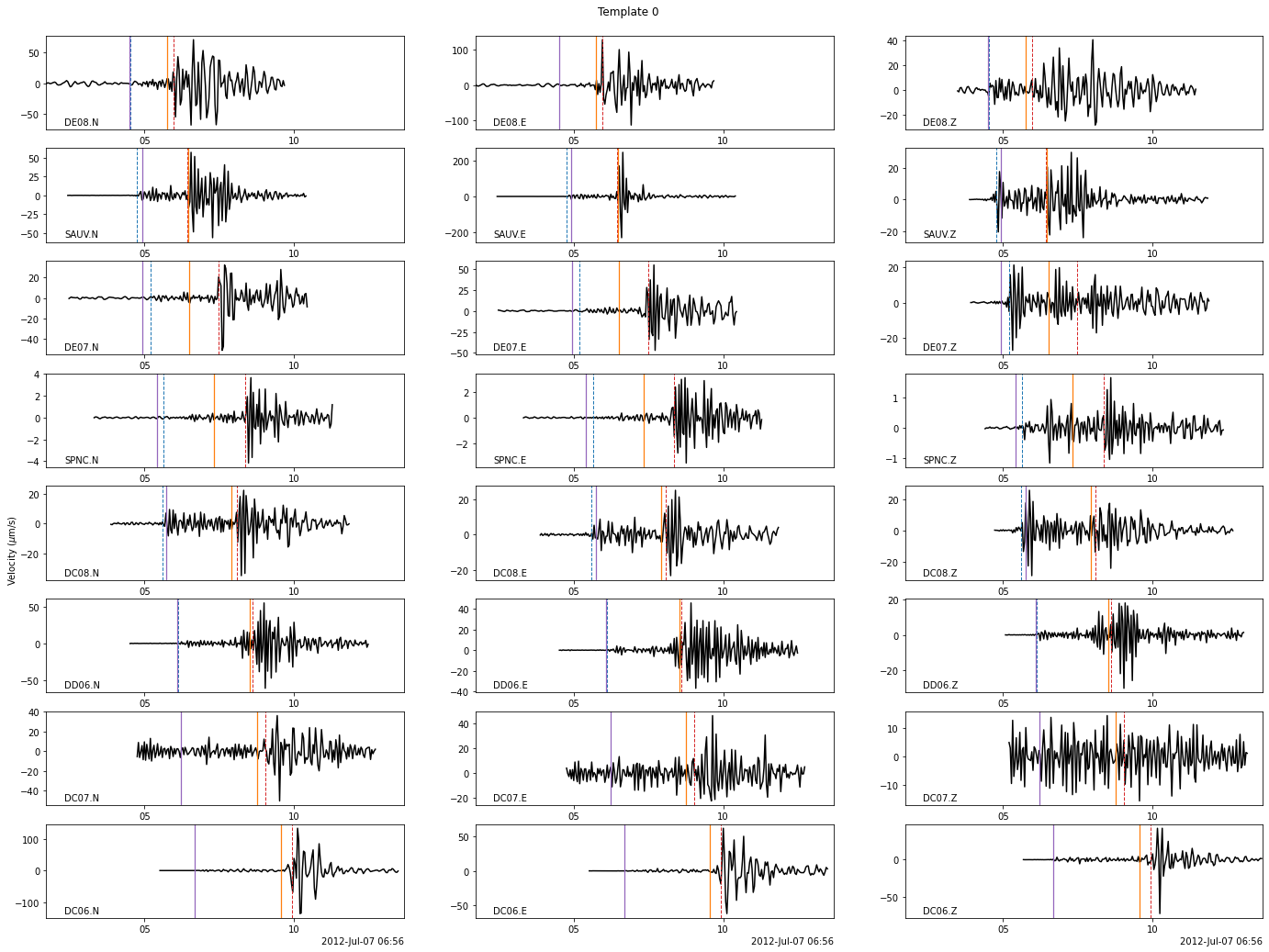
Let’s plot the waveforms of all the templates similar to template 0 on SAUV.E .
[24]:
from scipy.signal import hilbert
similar_tids = [0, 1, 3, 5, 9]
fig = plt.figure("waveforms_similar_templates", figsize=(15, 7))
ax = fig.add_subplot(111)
for tid in similar_tids:
tp = template_group.templates[template_group.tindexes.loc[tid]]
wav = tp.traces.select(station="SAUV", component="E")[0].data
norm = np.std(wav)
# align traces on max of envelope
time = np.arange(len(wav))
time -= np.abs(hilbert(wav)).argmax()
# time -= np.abs(wav).argmax()
if tid == 0:
ax.plot(time, wav/norm, color="k", lw=1.5)
else:
ax.plot(time, wav/norm, color="dimgrey", lw=0.75)
ax.set_xlabel("Time to max (samples)")
ax.set_ylabel("Velocity (AU)")
[24]:
Text(0, 0.5, 'Velocity (AU)')
The similarity of these templates would make them detect the same events and thus, spending time on redundant computation and writing redundant info in databases.
Save the non-redundant templates in a database
[25]:
output_dir = os.path.join(BPMF.cfg.OUTPUT_PATH, DB_PATH_T)
if not os.path.isdir(output_dir):
os.mkdir(output_dir)
for t, template in enumerate(templates_filtered):
tid = t
template.id = tid
template.set_aux_data({"tid": tid})
full_path = os.path.join(output_dir, f"template{tid}.h5")
template.write(
f"template{tid}.h5",
db_path=output_dir,
overwrite=True,
)